Interdisciplinary Graduate Track in Structural and Computational Biophysics (SCB)
Programs of Biology, Chemistry, Computer Science, Mathematics,
Molecular and Cellular Biosciences and Physics
Director, Freddie R. Salsbury Jr (salsbufr@wfu.edu)
This document provides a general overview of the requirements for students
participating in the SCB Track. For specific up-to date requirements,
please see the Wake Forest University Graduate Bulletin.
Track Overview: The Interdisciplinary Graduate Track in Structural
and Computational Biophysics (SCB) is designed to meet the need
for scientists and educators with broad, interdisciplinary training in the
quantitative biological, biochemical, and biomedical sciences. Students
who successfully complete the SCB Track and degree requirements
will receive a certificate in Structural and Computational
Biophysics, as well as the degree in the program in which they matriculate.
The Track is implemented by collaboration among the programs
of Biology, Chemistry, Computer Science, Mathematics, Molecular
and Cellular Biosciences and Physics at Wake Forest University.
Student Admission Requirements: Following matriculation
and at least one semester of coursework in a participating program
(currently, Biology, Chemistry, Computer Science, Mathematics,
Molecular and Cellular Biosciences and Physics), students can apply
for admission to the SCB Graduate Track. Admission to the
Track is initiated by meeting with the SCB program representative
or with the track directory. The student will then submit a letter of
intent and a Wake Forest University graduate transcript to their department
representative who will present it to the SCB advisory
committee. The letter of intent should express the student’s interest
in the SCB program, a proposed plan of study, and how the SCB
program meets the student’s career and academic goals. Following
favorable evaluation, applicants may be recommended for admis-
sion by the SCB advisory committee, with final approval determined
by the Graduate School. atics, molecular and cellular biosciences
or computer science).
Students in the Interdisciplinary Graduate SCB Track must
complete all graduate degree requirements in the individual program
to which they were admitted. In addition, at least 15 hours of
the student’s graduate coursework should consist of courses approved
as part of the SCB Track (listed in this bulletin), including a
general, introductory SCB course and two hours of journal club
credit. At least one course must be at the 700 level. Students must
take at least two graduate hours in each of the curriculum areas:
chemistry/biochemistry, computer science/mathematics, and biophysics.
All students in the SCB Track must complete and defend a
PhD dissertation (or MS thesis for computer science or mathematics)
that involves original, interdisciplinary research in the area of
structural and computational biophysics or computational biology;
broadly defined. The dissertation committee will consist of members
from at least three participating SCB departments. All students
must successfully complete a course in scientific ethics. Each semester,
several seminars from the participating departments will be
designated as SCB discussion group seminars. Students in the
Track are required to attend these seminars.
We encourage applications from underrepresented minorities and students with disabilities.
SCB Certificate Training Faculty
Rebecca Alexander (Chemistry):
Mechanisms of protein synthesis, aminoacyl-tRNA synthetases
Ed Allen (Mathematics): Combinatorial algorithms; computational
algebraic modelling; computational systems biology
Uli Bierbach (Chemistry):
Organic and inorganic synthesis of novel drugs and drug conjugates
Keith Bonin (Physics): Molecular
motors; total internal reflection fluorescence microscopy
James F. Curran (Biology): Molecular Mechanisms of Protein Synthesis, Gene Regulation, Nucleic Acid Chemistry, Microbial Evolutionary Mechanisms
Larry Daniel
(Biochemistry): Cell signaling in
cancer, lipid-derived messengers, redox networks;
modeling of signal transduction pathways
Jacquelyn Fetrow
(Physics and Computer Science): Protein structure and function relationships; protein motions and dynamics;
computational systems biology; biological network modeling.
Martin
Guthold (Physics): Nanobiotechnology , atomic force microscopy, fluorescence microscopy,
single molecule biophysics, fibrin fibers, blood clotting, aptamers
Adam Hall
(Biomedical Engineering): Single-molecule techniques and electical deduction of disease-relevant nucleic acid structural elements.
Tom Hollis
(Biochemistry): X-ray crystallography, DNA repair proteins, protein DNA interactions, processing nucleic acids to prevent autoimmune disease.
David John (Computer Science): Combinatorial
algorithms; genetic algorithms; computational algebraic modeling; computational
systems biology
Dany Kim-Shapiro (Physics): Spectroscopy, sickle cell hemoglobin, nitric oxide
Bruce King
(Chemistry): Organic compounds as nitric oxide (NO) and nitroxyl (HNO) delivery agents,
reagents to examine oxidative signaling pathways
Todd Lowther
(Biochemistry): Macromolecular X-ray crystallography, enzymatic reduction of cysteine sulfinic
acid and methionine sulfoxide, thioesterase and glyoxylate / hydroxypyruvate reductase structure / function
Doug Lyles(Biochemistry): Virus structure and assembly; biophysics and
genetics of viral protein function; signaling pathways in virus-infected cells
Jed Macosko (Physics): Mechanics of
protein machines and motors; atomic force microscopy (AFM), single molecule
fluorescence microscopy and video-enhanced differential interference contrast
light microscopy (VE-DIC)
Gloria Muday (Biology): Transport protein
structure/function, plasma membrane protein targeting, phosphorylation-dependent
signaling pathways
James Norris
(Mathematics): Statistical Methodology, Ecological Statistics
Derek Parsonage
(Biochemistry): Structure-function studies of the FAD-dependent
streptococcal NADH peroxidase and NADH oxidase; site-directed mutagenesis
V. Paul Pauca (Computer Science): Image processing.
Fred Perrino (Biochemistry): DNA replication, DNA
repair, mutagenesis, carcinogenesis, cancer therapeutics, deoxyribonucleases,
TREX genes, DNA polymerases, alkylation DNA damage, mechanistic and structural
studies
Leslie Poole
(Biochemistry): Enzymological and biophysical studies of antioxidant enzyme
systems; flavoprotein structure and function; redox active cysteinyl centers;
biological cysteine sulfenic acid formation
Fred Salsbury (Physics):
Molecular dynamics, protein dynamics and function, electrostatic and solvent models, structural modeling of proteins and
protein-DNA complexes, electronic structure
Peter Santago (Computer Science):
Pattern recognition, image and signal processing, machine learning, and high-performance cluster computing on various applications of pattern analysis.
Brian W. Tague (Biology):
Molecular genetic techniques to study plant cell biology and development.
Stan J. Thomas (Computer Science): Database theory, extracting information from data, and computer science education.
William Turkett (Computer Science):
Probabilistic reasoning, machine learning, bioinformatics, multiagent systems
SCB Curriculum Information
- Students will follow the curriculum for the Graduate Program in which they are seeking a degree (e.g. PhD in Biochemistry, MS in Computer Science). MS students must be pursuing thesis option.
- 15 hours in SCB-related courses including 2 hrs in each of three areas, 1 hour of discussion group for credit and 2 hours of journal club (the other 6 hours are in the student’s area of specialty). Coursework is deliberately flexible, and courses will be approved by program director.
- Students will successfully complete a course in scientific ethics (GRAD713/714 recommended).
- Student dissertation/thesis committee must have members from three different SCB associated departments.
- Students with at least one semester of coursework in a participating program can apply for
admission to the SCB Graduate Track.
A complete description of all classes can be found in the current graduate bulletin.
SCB-specific courses.
SCB 701. Structural and Computational Biophysics Journal Club.
(1) Seminal and current publications in structural and computational
biophysics are read and discussed. P—Admission to the SCB
graduate track or POI.
SCB 710. Research Topics in Structural and Computational Biophysics.
(1) Lectures and discussions on research topics in the field
of structural and computational biophysics and biology. Topics depend
on the specialty of the instructors in a given semester. P—
Admission to the SCB graduate track or POI.
Curriculum Area 1. Chemistry/Biochemistry
General prerequisites: Two semesters of undergraduate chemistry
and one semester of undergraduate biochemistry or molecular biology;
one semester of organic chemistry is considered ideal, but is
not required for most courses. (If additional prerequisites are required,
they are listed individually by course.)
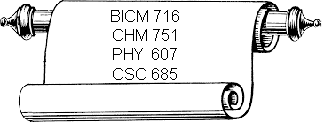
CHM 641. Fundamentals of Physical Chemistry. (3 or 4)
BICM 716. Special Topics in Biochemistry: Macromolecular Xray Crystallography. (2)
P—one semester graduate level biochemistry.
BIO 672. Molecular Biology. (3 or 4)
BIO/CHM 670. Biochemistry: Macromolecules and Metabolism. (3)
BIO/CHM 670L. Biochemistry Laboratory: Macromolecules and Metabolism. (1)
CHM 672. Biochemistry: Protein and Nucleic Acid Structure and Function. (3)
CHM 751. Biochemistry of Nucleic Acids. (3)
CHM 752. Protein Chemistry: Structures, Methods and Molecular Mechanisms. (3)
CHM 756. Biomolecular NMR. (1.5) P—POI.
CHM 757. Macromolecular Crystallography. (1.5) P—CHM 356A/656 highly recommended.
MCB 700. Analytical Skills (1) Taught every August.
MCB 701. Molecular and Cellular Biosciences A. (1) Taught every fall.
Curriculum Area 2. Physics
General prerequisites: Two semesters of undergraduate physics. (If
additional prerequisites are required, they are listed individually by
course.)
PHY 607. Biophysics. (3)
PHY 625. Biophysical Methods Laboratory. (1) C—PHY 607.
PHY 685. Bioinformatics. (3) P—Introductory courses in biology, chemistry, and molecular
biology or biochemistry or permission of instructor; also listed as CSC 685, though
requirements and prerequisites are different.
PHY 620. Physics of Biological Macromolecules. (3) P—PHY 651 or CHM 641, or POI.
Curriculum Area 3. Computer Science/Mathematics
General computer science prerequisites: Programming in a high level language. (If
additional prerequisites are required, they are listed individually by course.)
CSC 621. Database Management Systems. (3)
CSC 631. Object-oriented Software Engineering. (3)
CSC 646. Parallel Computation. (3)
CSC 652. Numerical Linear Algebra. (3)
CSC 655. Introduction to Numerical Methods. (3)
CSC 671. Artificial Intelligence. (3)
CSC 685. Bioinformatics. (3)
CSC 721. Theory of Algorithms. (3)
CSC 753. Nonlinear Optimization. (3) P—Computer Science 655.
CSC 754. Numerical Methods for Partial Differential Equations. (3) P—CSC 655 or
MTH 655.
MTH 652. Partial Differential Equations. (3) P—MTH 251.
MTH 653. Mathematical Models. (3)
MTH 656. Statistical Methods. (3)
MTH 659. Multivariate Statistics. (3) P—MTH 656 and 602.
MTH 750. Dynamical Systems. (3) P—MTH 611.
MTH 761. Stochastic Processes. (3)
T32-GM095440 Training Program in Structural and Computational Biophysics
Mission: To bridge the gap between biomedical sciences and more basic quantitative sciences
Our NIH T32 supported Structural and Computational Biophysics (SCB) training program is designed to bring students from diverse
educational backgrounds to the point where they speak a common scientific language. The students first achieve a level of
competency in their original discipline. This is provided by the discipline-specific coursework prescribed for each program.
After this phase, students work to bridge the gap between the disciplines. The SCB Training Program has developed four mechanisms
to achieve this instruction: advanced coursework in structural and computational biophysics,
the SCB Discussion Group,
Center for Structural Biology Seminar Series, and SCB Training Program retreats.
All four of these mechanisms bring together students from the biomedical programs and Chemistry and Physics with the
goal of developing interdisciplinary training.
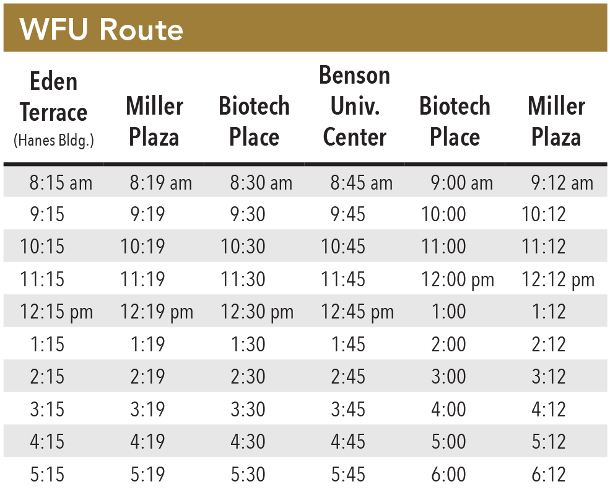
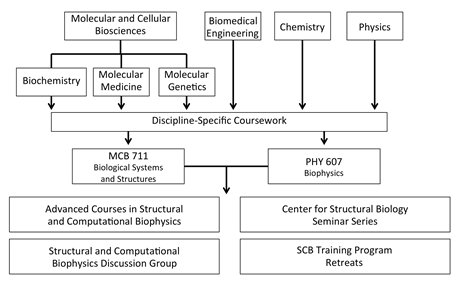 Curriculum organization for students in the SCB Training Program.
Curriculum organization for students in the SCB Training Program.
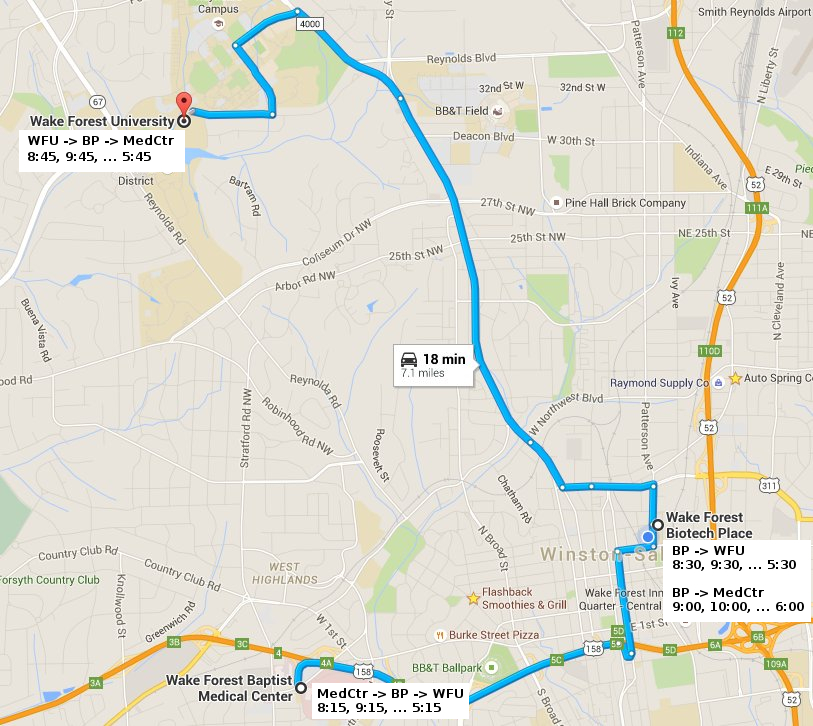
 2015 SCB retreat at Biotech Place.
2015 SCB retreat at Biotech Place.
The members of the core training faculty belong to the departments of Biochemistry, Biomedical Engineering, Chemistry, Molecular Medicine,
and Physics. Additional associated training faculty come from the departments of Biology, Computer Science, and
Mathematics. Many of the faculty have significant research interactions with each other and work to answer questions
in the areas of molecular signaling, redox biology, and DNA damage and repair.
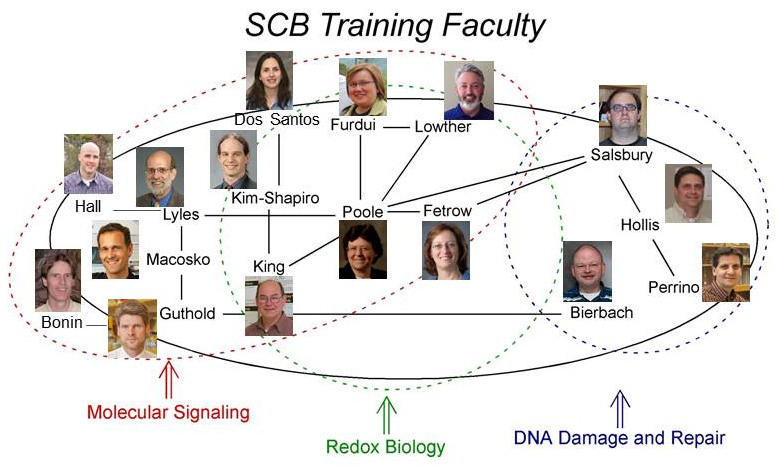
Click on faculty photos above for link to research websites.
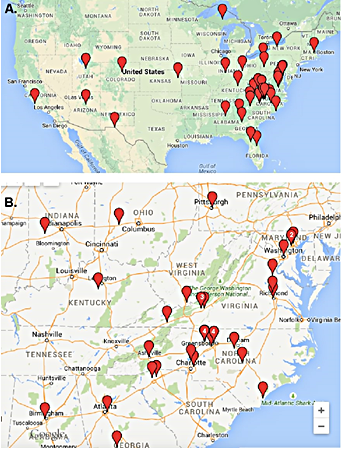
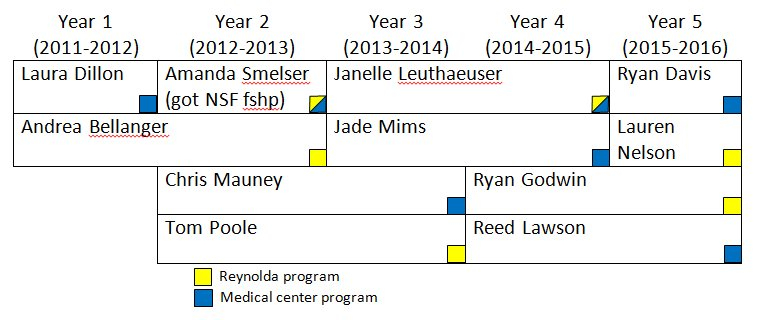
Students supported by the T32 Training Grant.
Application
Application Process (Graduate
School applicants): For those applying for acceptance into graduate school, application is made to the home program (Biology, Chemistry, Computer Science, Mathematics, Molecular and Cellular Biosciences or Physics.
You are encouraged to express, at the time of application, your interest in
being considered for the"SCB Track" at the beginning of your "Statement
of Interest".
Submit the WFU
graduate school application through the Graduate School website.
Formal Admission to the
SCB Track: Students are considered for admission to the Interdisciplinary
SCB Graduate Track after the first semester following their admission to the
graduate program in the department to which they initially applied.
Admission to the Track is initiated by meeting with the SCB
department representative. The student will then submit a letter of intent
and a Wake Forest University graduate transcript to their department representative,
who will submit it to the SCB Advisory Committee. The letter of intent should
express the student's interest in the SCB program, a proposed plan of study,
and how the SCB program meets the student's career and academic goals. Admission
is recommended by the SCB Advisory Committee; final admission to the SCB Track
is determined by the Graduate School.
We encourage applications from underrepresented minorities and students with disabilities.
Download Application Form
Contact Information:
(please direct your inquiries to the individual
listed below affiliated with the department of primary research interest)
Director: Fred Salsbury (Physics),
336-758-4975
336-758-4975
Departmental Representatives on the SCB Advisory Board:
Uli Bierbach (Chemistry),
336-758-3507
336-758-3507
Daniel Kim-Shapiro (Physics),
336-758-4993
336-758-4993
Gloria Muday (Biology),
336-758-5316
336-758-5316
James Norris (Mathematics),
336-758-4888
336-758-4888
Leslie Poole (Biochemistry),
336-716-6711
336-716-6711
William Turkett (Computer Science),
336-758-4427
336-758-4427
Mailing Address for: Salsbury, Allen, Bierbach, Kim-Shapiro, Muday and Turkett:
Wake Forest University
Reynolda Station
Winston-Salem NC 27109
Mailing Address for Poole:
Wake Forest University School of Medicine
Medical Center Blvd
Winston-Salem NC 27157